Protein Kinase Ontology
| Current version: 67 Date: 2023-10-11
The conceptual representation of such diverse information in one place, in the form of the ProKinO knowledge graph, enables not only rapid discovery of significant information related to a specific protein kinase, but also enables large scale integrative analysis of the protein kinase family. An outline of the data organization in ProKinO is represented using a UML class diagram (see Schema, below). Publications: Soleymani, S., Gravel, N., Huang, L. C., Yeung, W., Bozorgi, E., Bendzunas, N. G., Kochut, K.J., and Kannan, N. (2022). Dark kinase annotation, mining and visualization using the Protein Kinase Ontology. bioRxiv, 2022-02. doi: https://doi.org/10.1101/2022.02.25.482021 Salcedo, M., Gravel, N., Keshavarzi, A., Huang, L., Kochut, K., Kannan, N. (2023). Predicting Protein and Pathway Associations for Understudied Dark Kinases using Pattern-constrained Knowledge Graph Embedding. PeerJ, under review Yeung, W., Zhou, Z., Li, S., & Kannan, N. (2023). Alignment-free estimation of sequence conservation for identifying functional sites using protein sequence embeddings. Briefings in Bioinformatics, 24(1), bbac599. McSkimming, D.I., Dastgheib, S., Talevich, E., Narayanan, A., Katiyar, S., Taylor, S.S., Kochut, K.J., Kannan, N. (2015). "ProKinO: a unified resource for mining the cancer kinome." Human Mutation 36(2):175-86. doi:10.1002/humu.22726. Gosal, G., Kochut, K.J., Kannan, N. (2011). "ProKinO: An Ontology for Integrative Analysis of Protein Kinases in Cancer." PLoS ONE 6(12):e28782. doi:10.1371/journal.pone.0028782 Gosal, G., Kannan, N., Kochut, K.J. (2011). "ProKinO: A Framework for Protein Kinase Ontology," in Proceedings of the 2011 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 550-555. |
Data Sources
The following table shows the versions of the datasources used for population of the ProKinO version loaded in this browser.
Disease Data COSMIC (Catalogue of Somatic Mutations in Cancer) database ` is used as the source of information regarding kinase cancer mutations. In addition to mutations, other information such as, the primary sites, histology, samples, description and other relevant features is also obtained from COSMIC. The mutation data was obtained from the Sanger Institute Catalogue Of Somatic Mutations In Cancer web site, http://www.sanger.ac.uk/cosmic Bamford et al (2004) The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer, 91,355-358. Pathway and Reaction Data The pathway and reaction data in ProKinO is obtained from Reactome.Reactome project. http://www.reactome.org/ (26th January 2011). Function Data The information regarding functional domains and functional features associated with kinase domains is obtained from UniProt. The information regarding the functional domains associated with kinase domains, the crystal structures solved for each kinase domain, isoforms, and functional features associated with kinases, such as the modified residue, signal peptide, topological domain, cellular location and tissue specificity, are obtained from UniProt.The UniProt Consortium, Ongoing and future developments at the Universal Protein Resource Nucleic Acids Res. 39: D214-D219 (2011). Classification and Sequence Data Data regarding protein kinase sequence and classification is obtained from KinBase.The Protein Kinase Complement of the Human Genome, G Manning, DB Whyte, R Martinez, T Hunter, S Sudarsanam (2002). Science 298:1912-1934. Pseudokinase data and their classification Pseudokinase sequences were identified from the Uniprot reference proteomes database and classified into pseudokinase families in the Kannan Lab at the University of Georgia. Pseudokinase protein names were either obtained from UniProt or assigned by the pseudokinase classification if the protein name is not provided by UniProt.Kinase Sub-domain Data To capture the sub-domain information in ProKinO, we have used a motif model [1, 2] with key motifs corresponding to each of the sub-domains in the kinase domain [3].
|
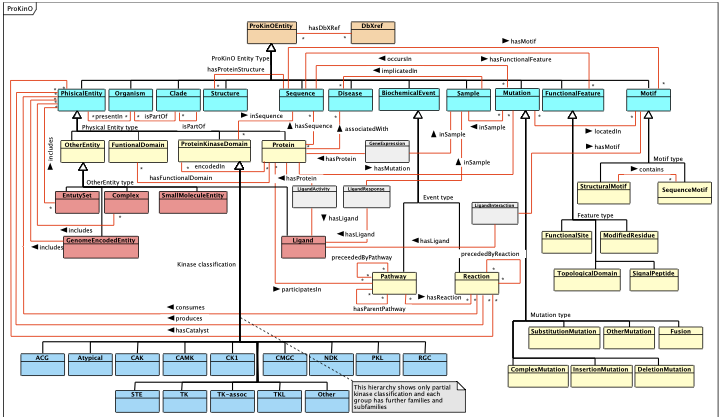
ProKinO Schema
| The picture below is an outline of the ProKinO schema as a UML class diagram. It shows major ProKinO classes and object properties (relationships connecting ProKinO classes). It is available in PDF, as well |
ProKinO Browser
You can use this ontology browser to quickly locate protein kinase proteins and a lot of information related to the proteins, including the sequence, structure, function, mutation and pathway information on kinases. You may initiate ProKinO browsing by selecting one of the following items in the Browse menu:
It is also possible to initiate ProKinO browsing by searching for a specific kinase protein, disease, pathway, or for any object. The searching uses a set of terms, possibly separated by OR, AND and AND NOT boolean connectors. It is possible to use wildcards, for example, FGFR*. Phrases should be enclosed within double quotes. The Query functionality for Proteins, Arbitrary SPARQL queries can be submitted (however, the size of the resulting set is limited to 1500, at this time). The ProKinO ontology has not been released for downloading, yet. After you initiate ProKinO browsing by selecting and displaying the data information page for one of the proteins, kinase domains, diseases, or functional domains, you will be able to explore the ontology by activating the hyper-links leading to a variety of related data. Usually, a data information page includes the properties of the shown entity (for example, cellular location or tissue specificity of the proteins), and links to other related ontology entities (for example, functional domains, sequences, pathways of the proteins). The protein data is subdivided into sub-areas, including the general information, functional features, mutations, pathways, and external references. These sub-areas of information are available by clicking on the provided tabs. The mutations are further subdivided into substitutions, insertions, deletions, complex mutations, and other. |
People
ProKinO is a collaborative effort between the Evolutionary Systems Biology Group Lab of Dr. Natarajan Kannan at the Biochemistry and Molecular Biology Department and Dr. Krys J. Kochut's lab at the Computer Science Department, both at University of Georgia, Athens, USA. Gurinder Gosal, also at UGA, created the initial version of the software system to automatically populate ProKinO from the selected data sources.
Current Team Members

Dr. Natarajan Kannan
BioInformatics

Dr. Krzysztof Kochut
Computer Science

Dr. Saber Soleymani (CS)
Georgia State University

Nathan Gravel
Bioinformatics

George Bendzunas
Bioinformatics

Elika Bozorgi
Computer Science

Amitabh Priyadarshi
Computer Science

Safal Shrestha
BioInformatics
Former Team Members

Dr. Liang-Chin Huang (IOB)
Melax Tech

Dr. Abbas Keshavarzi (CS)

Dr. Annie Kwon (IoB)
UCSF

Dr. Daniel McSkimming (IoB)
Comp. Biology, USF

Dr. Shima Dastgheib (CS)
Genentech/Roche

Dr. Gurinder Gosal (CS)
British Columbia

Reshmi De (CS)
Morgan Stanley

Anish Narayanan, M.D. (IoB)
UTHSC-San Antonio

Bhargabi Chakrabarti (CS)
George Mason Univ.